Genetic Mutations Conferring Resistance to Candida albicans to Antifungal drugs: A Global Perspective and Regional Implications
PM Sawadogo1,2, A Zida1,2, I Sangaré5,6, TK Guiguemdé2,3, A. Sanfo1, M. Idani1, H Nacanabo1, S Bamba5,6, R Ouédraogo/Traoré2,3, TR Guiguemdé2,4
1Parasitology-Mycology Department, Yalgado Ouedraogo University Hospital Center, Ouagadougou, Burkina Faso
2Training and Research Unit in Health Sciences, Ouaga University 1 Professor Joseph Ki-Zerbo (UO1 / PrZKZ), Ouagadougou, Burkina Faso
3Parasitology-Mycology Department, Charles de Gaulle University Hospital Center, sector 28 Ouagadougou, Burkina Faso
4Muraz Research Center, Bobo-Dioulasso, Burkina Faso
5Parasitology-Mycology Department, Souro Sanou University Hospital, Bobo Dioulasso, Burkina Faso
6Institut de Reccherche en Sciences de la Santé, Université Nazi Boni (UNB), Bobo Diuolasssa, Burkina Faso
Abstract
This article aims to summarize the results of works from January 2013 to December 2017, on the molecular mechanisms of Candida albicans resistance to antifungal drugs. It is a prelude to a study on the molecular mechanisms of these resistances in Burkina Faso, with the aim of exploring new therapeutic solutions. Almost all studies have focused on the ERG11 gene as the most involved in azoles resistance. Mutations have also been demonstrated on other genes conferring resistance to other molecules such as CDR1 and 2, MRR1 and 2, TAC1 and ERG for polyenes, allylamines and azoles, FKS1 for echinocandins, FCA1 and FCY1 for pyrimidine analogues. Genetic mutations conferring the resistance of C. albicans to antifungals drugs worldwide are regularly reported, but in Burkina Faso we have no data on this subject. As a perspective therefore, a study on the molecular mechanisms of resistance of C. albicans to antifungals will be of great help in the fight against the resistance of this frequent yeast to antifungals drugs.
Introduction
In Burkina Faso, candidiosis is a real public health problem. Candida albicans is involved in more than 80% of these candidiosis whose spectrum ranges from vaginal, oral, cutaneous candidiosis to deep candidiosis1(figure 1 and 2). Thus, to combat these infections, antifungals are used, the misuse of which has led to the emergence of C. albicans resistance to these molecules2,3,4,5. For example, fluconazole used as a preventive treatment for people living with Human immuno-deficiency virus (HIV) in Burkina Faso experienced a 66.5% decrease in susceptibility to C. albicans6. In the literature, several studies deal with the molecular mechanisms of C. albicans resistance to antifungals3,6,7,8,9,10,11,12,13. In our context in Burkina Faso, the studies carried out so far are limited to the search for susceptibility profiles of C. albicans to antifungals and the identification of the genotypes involved6. However, it is necessary to elucidate these molecular mechanisms in our country to explore new therapeutic solutions. This review is therefore a prelude to work on the molecular mechanisms of C. albicans resistance to antifungal drugs in Burkina Faso. It allows us to take stock from literature data, studies of the past five years on these resistance mechanisms elsewhere in the world in order to have a clear idea of the importance of the problem.
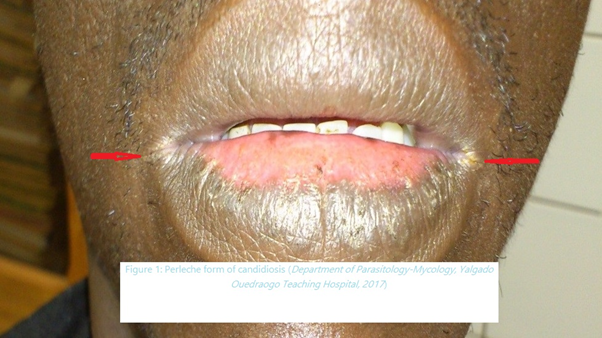
Figure 1: Perleche form of candidiosis (Department of Parasitology-Mycology, Yalgado Ouedraogo Teaching Hospital, 2017)
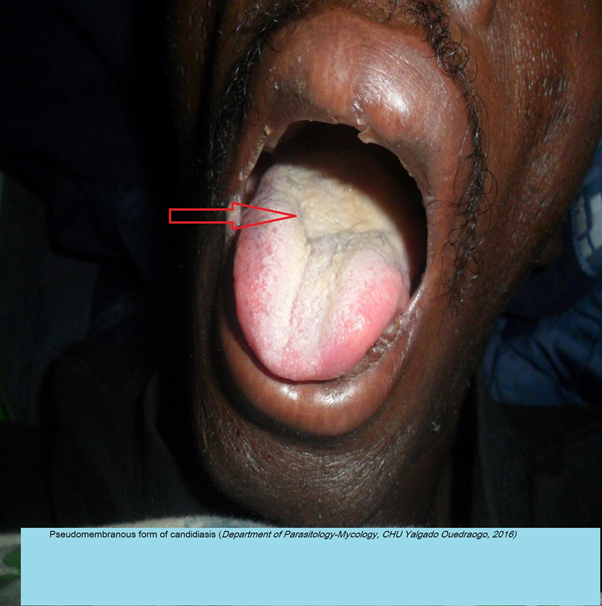
Figure 2: Pseudomembranous form of candidiasis (Department of Parasitology-Mycology, CHU Yalgado Ouedraogo, 2016)
Data Collecting
This study was conducted in Burkina Faso
This is a review of studies that examined the molecular mechanisms of C. albicans resistance to antifungals in the last five years, from 2013 to 2017. The articles were collected from January 2018 to February 2018.
The pubmed database served as a research base. As for the other bases Hinari and google scholar, we consulted them to have access to the full text of some articles.
For the translation of the terms into English, we used the CISMEF website (catalog and index of French-language medical sites: http://www.chu-rouen.fr/cismef/) which allowed us to find the words- key.
The search for the articles was done with great care using the following search terms: Genetic mutations / genomic instability /Candida albicans / antifungal drugs resistance. The selection of the studies to be included required the prior establishment of inclusion and non-inclusion criteria.
The articles that met the following criteria were used to conduct the review: studies that demonstrated the C. albicans strains resistance to antifungal drugs and the molecular mechanisms underlying this resistance; studies written in French or English; studies conducted between 2013 and 2017.
The following articles were not included: articles that focused solely on antifungal susceptibility studies; studies dealing with combinations of antifungals and other molecules; studies that used non-antifungal molecules.
For any selected article, we collected the following data: name of the principal author, date of publication, types of strains (clinical strains, laboratory strains), the molecules used, the resistance genes and their mutations.
A total of 126 articles were included in the review.
Strains of C. albicans Used in the Different Studies
The C. albicans strains used were essentially clinical strains and incidentally laboratory strains as controls (Table 1)3,14,15,16,17. For example, Caban et al. in 2016 used three laboratory strains called Ca1r, Ca11r, and Ca12r, as control strains18.
Table 1: Genes studied, strains used and genetic mutations of Candida albicans
| Genes or groups of genes studied | Strains used | molecules used | Main mutations of resistance and overexpression of genes or group of genes | sources |
|---|---|---|---|---|
| ERG11 | Clinica | FCZ | E116D F145L I437V | Arati M et al. 2016 |
| Clinical | azoles (FCZ, ITZ et VCZ), | A530C, G622A, G1309A, A1167G, A1230G. | Katarzyna J et al. 2013 | |
| clinical + laboratory | azoles (FCZ, KTZ, MCZ, ITR), AMB, 5FC | Q266AspD, L480Q | Caban M et al.2016 | |
| Clinical | azoles (FCZ, VCZ) | A114S, Y132H, Y132F, K143R, Y257H K143Q | Xiang MJ et al. 2013 | |
| Clinical | FCZ | D225H, K342R, G450E V488I G129A Y132H, A114S, Y257H, V437I, G465S, G448E et K128T | Ying Y et al. 2013 | |
| Clinical | FCZ | A61V, D116E, K128T, Y132F, E266D, S279F, L321F, S405F, V488I, S405F | Carvalho VO et al. 2014. | |
| clinical | Y132H et G450E, Y257H G464S, | Lei Z et al. 2013 | ||
| CDR1,CDR2, MDR1,ERG11 | clinical | FCZ | overexpression of CD1 gene | Salari S et al. 2015 |
| ERG11,CDR1, MDR1, Flu1 | clinical + laboratory | FCZ | ERG11p mutations (A114S and Y257H) + overexpression CDR1, MDR1, and Flu1 | Xu Y et al. 2015 |
| ERG11,CDR1, CRD2, MDR2 | clinical | azoles (FCZ, ITZ, VCZ) | overexpression of CDR2, MDR1 and ERG11 + ERG11p mutations (D116E, K128T, V159I and E266D), | Go??bek K et al. 2015 |
| FKS1 | Clinical | echinocandins (CFG, MCF) | S645P R1361H S645F | Dudiuk C et al. 2015 |
| FKS1, ERG11, ERG2, CDR1 CDR2, TAC1 | Clinical | Azoles, and echinocandins | Erg11 mutations (E266D, G307S, G450E and V488I) + Tac1 mutations (R688Q) + Fks1 mutation (S645P) + increased overexpression of ERG11 and CDR2 | Jensen RH et al. 2015 |
| ERG11, ERG3, MDR1, MRR1 | clinical + laboratory | azoles (FCZ , VCZ), polyenes (AMB) | mutation MRR1, ERG11 and ERG3 + overexpression Mdr1, MRR1, | Eddouzi J et al. 2013 |
| ERG11, MDR1, TAC1, CDR1, CDR2 | Clinical | azoles (FCZ et VCZ), allylamines | ERG11 mutations (A114S, Y132H, Y132F, K143Q, K143R, Y257H and G448E) and TAC1 (F964Y) | Liu JY et al. 2015 |
| ERG11, ERG4 | Clinical | 5FC, AMB, FCZ, ITZ, VCZ | 2 silent mutations on ERG4 and 10 mutations on ERG11 | Feng W et al. 2017 |
| TAC1, CDR1, CDR2 | Clinical | azoles | Mutations on TAC1 + overexpression of CDR1 and CRR2 genes | Liu Z et al. 2017, Popp C 2017 |
| ERG11, TAC1, MRR1, UPC2, FKS1 | Clinical | Azoles et echinocandins | 126 substitutions: TAC1 (38) ERG11 (20) UPC2 (15) MRR1 (33) | Sitterlé E et al. 2017 |
| FUR1, FCA1, FCY1 | Mutations FUR1 (R101C) + mutations FCA1, FCY1 | Arendrup MC et al. 2017 |
FCZ (flconazole), ITZ (itraconazole), VCZ (voriconazole), KTZ (ketoconazole), MCZ (miconazole), AMB (amphtecin B), CFG (caspofungin), MCF (micafungin), 5FC (5flucytosine)
To better correlate mutations and resistance to antifungal drugs, many authors have used two types of clinical strains in their studies: sensitive strains and resistant strains19. And Some authors have preferred to confront three types of strains: sensitive clinical strains, resistant clinical strains and laboratory strains20.
From a general point of view, we find that most of the strains used in the different studies were clinical strains. The explanation is that these strains would be more available and accessible compared to laboratory strains. Moreover, the results from the manipulation of these strains would be closer to reality, these strains coming directly from sick people in whom the resistances were observed.
In Burkina Faso, studies on the resistance of C. albicans to antifungals have so far focused on two aspects of the phenomenon: on the one hand, the phenotypic study of the in vitro sensitivity of clinical strains of C. albicans to antifungals and other hand genotypic study of the sensitivity of clinical strains of C. albicans6. These studies revealed a high frequency of azole-resistant strains, notably fluconazole (resistance observed with 70% of the strains studied)6. Of the genotypes A, B and C identified, the genotype A is the most common and most involved in antifungal resistance6. However, no studies have yet been conducted on the resistance genes of C. albicans in Burkina Faso.
Antifungal Drugs Used in the Different Studies
The majority of authors have focused on resistance to azoles, particularly fluconazole (Table 1)6,11,14,21,22. Sometimes other azoles have been associated with fluconazole to demonstrate cross-resistance to azoles. These are voriconazole Iitraconazole, ketoconazole and miconazole9,16,17,18,19,23,24,25,26. Indeed, fluconazole is the molecule most used to treat candidosis. The clinical response is generally good, but because of its fungistatic effect, relapses are common. Thus, prolonged and repeated treatment promotes the emergence of resistance to azoles in general and fluconazole in particular19,27,28.
Nevertheless, the literature reports studies on the C. albicans resistance to other chemical groups but to a lesser extent: echinocandins29, polyenes18,23,24, allylamines candidiosis23,30.
Mechanisms of Resistance to Antifungals
To azoles
The mechanisms of resistance to azoles are varied and can be divided into four major groups:
Modification of the target (14-alpha-demethylase)
The modifications of the 14-alpha-demethylase enzyme correspond to amino acid substitutions in the protein sequence31,32. These modifications result in decreased affinity of 14-alpha-demethylase for azoles or conformational changes preventing access of the antifungal agent to the active site31. The azoles cannot then play their inhibitor role. These substitutions are the consequence of mutations point in the ERG11 gene coding for 14-alpha-demethylase33,34. Many different mutations have been described.
The modifications of the 14-alpha-demethylase enzyme correspond to amino acid substitutions in the protein sequence31,32. These modifications result in decreased affinity of 14-alpha-demethylase for azoles or conformational changes preventing access of the antifungal agent to the active site31. The azoles cannot then play their inhibitor role. These substitutions are the consequence of mutations point in the ERG11 gene coding for 14-alpha-demethylase33,34. Many different mutations have been described.
According to Carvalho et al. in 2014, more than 140 resistant mutations were described on the C. albicans ERG11 gene21. As such, many authors have been interested in this gene in studies of the C. albicans resistance to antifungals. The frequencies of the mutant strains in relation to the ERG11 gene vary from one study to another. Arati et al.14 in 2016 found 34% mutations and 48.1% mutant strains were resistant to fluconazole. Zhang et al. in 2013 and Ying et al. in 2013 found respectively 41% mutant strains with azoles and 57.9% mutant strains with fluconazole22,35.
All reported mutations are nucleotide substitutions generating or not (silent mutations) a change of amino acids at a level of the encoded protein (ergosterol). Xiang et al.17 in 2013 showed that ERG11p (14α-demethylase) substitutions A114S, Y132H, Y132F, K143R, Y257H and K143Q were associated with resistance to fluconazole and voriconazole. Arati et al. in 2016, found two resistant ERG11p mutations: E116D and F145L14. Katarzyna et al.16 in 2013 highlighted the following mutations on the ERG11 gene: A530C, G622A, G1309A, A1167G, and A1230G. These mutations were observed on resistant strains. But having not obtained any significant association, they then concluded that an isolated mutation on the ERG11 gene does not necessarily affect the sensitivity of C. albicans to azoles, whereas multiple nucleotide substitutions on the ERG11 gene would affect the sensitivity of C. albicans to azoles16. Xiang et al.17 in 2013 had reached the same conclusion.
In addition, in this study, a new ERG11p K143Q mutation, which has never been demonstrated by previous studies, has been described in resistant strains17. Ying et al.22 in 2013, showed the following ERG11p mutations in fluconazole-resistant strains. Y132H, A114S, Y257H, V437I, G465S, G448E, and K128T: And like the above-mentioned authors they had reached the same conclusion that these multiple amino acid substitutions in Erg11p were frequently found in resistant strains and could be associated resistance to fluconazole22,36,37. Carvalho et al.21 in 2014 in a letter to the publisher claimed that the mutation ERG11p S405F would be responsible for an increase of four times the minimum inhibition concentration (MIC) of fluconazole against C. albicans. However, mutations of the ERG11 gene are only one of the mechanisms that may lead to resistance to fluconazole and should not be considered in isolation21,38.
In conclusion, the authors are unanimous on the following remarks: one substitution observed on the ERG11 gene in mutant strains of C. albicans alone would not be sufficient to confer acquired resistance to azoles, because of the ERG11 gene polymorphism23,35. Thus some mutations are not responsible for resistance azoles23. Mutations that cause microbiological resistance are grouped in three very specific regions of the gene azoles23,35.
Other ERG genes such as ERG1, ERG3, ERG6, ERG24, ERG25 are also involved in the C. albicans resistance to azoles16,23. However, mutations on these genes have been rarely reported23.
Overproduction of the target (14-alpha-demethylase)
By increasing the production of the target enzyme, C.albicans can decrease its sensitivity to the activity of azole antifungals18,34,39. Nevertheless, this mechanism must be associated with other mechanisms to achieve resistance in clinical strains18,34,40. The overproduction of 14-alpha-demethylase may be the result of two mechanisms. On the one hand, it may be an increase in the expression of the ERG11 gene, and on the other hand, the overproduction may be the consequence of an increase in the number of copies of the ERG11 gene30,40,41. The regulation of the transcription of the ERG11 gene in C. albicans is complex40. The transcription factor UPC2 makes it possible to control the level of expression of certain genes including ERG11. Mutations "gain of function" of UPC2 can be at the origin of an increase of the activity of this gene, which will result in overexpression of the ERG11 gene antifungals20, 32, 36,41. This type of resistance mechanism has been demonstrated in C. albicans strains of clinical origin. Sitterlé et al. in a panoramic study of resistance genes on 151 strains of C. albicans to antifungal drugs in 2017 made the following conclusion: a total of 126 substitutions identified ,15 concerned UPC2 gene11. Mutations in this gene were associated with resistance to fluconazole11.
The overproduction of 14-alpha-demethylase may also be related to an increase in the number of copies of the ERG11 gene occurring as a result of chromosomal rearrangements9,41,42,43,44.
Phenomena of efflux
To exercise their antifungal activity, azoles must enter the fungal cell and be at an intracellular concentration sufficient to inhibit 14-alpha-demethylase39. C. albicans has, naturally, at the level of their plasma membrane, multidrug carriers allowing the efflux of different molecules39. The CDR1, CDR2 and MDR1 genes encode efflux pumps that are membrane transporters that excrete toxic molecules out of the fungal cell. CDR1 and CDR2 encode ABC transporters, and the MDR1 gene encodes MFS transporters24,39.
Increased expression of these transporters (efflux pumps) is an important mechanism of azole resistance in clinical C. albicans strains39. In recent years, numerous studies have demonstrated the importance of transcriptional regulation of the genes encoding these transporters in the acquisition of azole resistance.
According to Popp et al. in 2017 and Zhang et al. in 2017, overexpression of these genes could help increase the activity of these pumps and cause resistance to azoles. This increase is the result of mutations in transcription factors such as, TAC1 and MRR1 which respectively control the expression of CDR and MDR1 genes24,40. In the literature there are cases of resistance to azoles following the overexpression of these genes. Thus, according to Salari et al. in 2015, overexpression of the CDR1 gene is strongly correlated with increasing resistance to fluconazole36. Other authors such as Go??bek et al. as well as Liu et al. reported overexpression of these genes (CDR1, CDR2 and MDR2) that encode efflux pumps was observed in resistant strains of C. albicans to azoles confirming the multifactorial character of resistance mechanisms of C. albicans already mentioned by some authors19,25.
In 2015 Jensen et al. reported that one or more mutations on the TAC1 gene would lead to overexpression of the CDR1, CDR2 and MDR2 genes and thus confer resistance to azoles. Rather in 2013 Eddouzi et al. 2013 demonstrated overexpression of the gene superfamily (CDR1, CDR2, MDR1, MDR2, and MRR1) coding for efflux pumps and strongly associated with resistance of c. albicans to azoles24.
Alterations of the ergosterol synthesis route
In the presence of azoles, the inhibition of 14-alpha-demethylase leads to the accumulation of methylated sterols which are transformed into toxic products by delta-5,6-desaturase, an enzyme which is encoded by the ERG3 gene19,25. If the ERG3 gene is mutated, these toxic products are no longer synthesized and the fungal cell can survive and thus become resistant to azoles19. This mechanism of resistance, although infrequent, has been demonstrated in some clinical strains of C. albicans19.
To polyenes
The mechanism involved could be related to a disappearance of ergosterol, the target of the antifungal45. This disappearance may be the consequence of a blockage of the ergosterol biosynthesis pathway by mutation of a gene that must be accompanied by the establishment of an accessory metabolic pathway allowing the synthesis of other essential membrane sterols to the survival of the fungal cell3,12,15,45. Thus, it has been shown that mutations in delta-5,6-desaturase encoded by the ERG3 gene were responsible for resistance to amphotericin B in clinically relevant strains of C. albicans45. In strains with this type of mutation, there is cross-resistance to fluconazole and for some strains a decrease in virulence45.
To echinocandins
The acquired resistance of C. albicans to echinocandins was initially studied in laboratory mutants11,15. It is only more recently that cases of resistance have been reported in C. albicans strains that cause infections in patients11,25. Resistance was initially described for caspofungin but is also present for other echinocandins (micafungin and anidulafungin)25. The three echinocandins having the same mechanism of action, it is a class resistance25.
Currently the only proven mechanism of acquired echinocandin resistance in C. albicans is the presence of mutations in the FKS genes that encode the target of this antifungal class, beta-1-3-D-glucan synthase29,30. At the level of the FKS gene, the mutations responsible for resistance are confined to two short sequences that each encode nine amino acids15. These regions have been called "hot spot" 1 (HS1) and "hot spot" 2 (HS2)15. Mutations are most commonly found in HS1 of FKS1 for C. albicans20. For example, The S645P mutation of the Fks1p gene is significantly associated with the resistance of C. albicans to echinocandins15. Depending on the position of the mutation in the hot spot, the level of resistance may be higher or lower15. Biochemical analyzes of beta-1-3-D-glucan synthase extracted from mutated resistant strains showed that the affinity of the enzyme for echinocandins was decreased, confirming the role of mutations in the occurrence of microbiological resistance15,20,30. The analysis of a large number of resistant strains shows that the microbiological resistance is therefore undergone by genetic alterations and biochemical modifications at the level of the target enzyme. These resistances have also been confirmed in animal models and are correlated with therapeutic failures in patients15.
To fluoropyrimidines (5-Fluorocytosine)
The acquired resistance to 5-fluorocytosine can appear rapidly during a treatment of a Candida infection, which does not allow to use this molecule as monotherapy28. As a result, 5-fluorocytosine is indicated only in combination with another antifungal agent, most commonly amphotericin B28. Different molecular mechanisms can cause resistance to 5-fluorocytosine28. Mutations on the FCA1, FCY1 and FUR1 genes would lead to resistance to 5-fluorocytosine. Indeed, FCA1 and FCY1 encode cytosine desaminase, and mutations in these genes therefore inhibit the conversion of flucytosine to 5-F-fluorouridine28.
FUR1 codes for uracil phosphoribosyltransferase, and mutations in this gene inhibit the conversion of 5-fluorouridine to 5-fluorodeoxyuridylic acid monophosphate30. And the substitution R101C on FUR1 is much associated with this resistance41,45.
Conclusion
Progresses in molecular biology have allowed researchers to better understand the molecular mechanisms of C. albicans resistance to antifungal drugs. The resistance of C.albicans to antifungals involves a variety of genes and mechanisms.
In Burkina Faso, resistance to fluconazole is the most observed. But no resistance genes yet studied. To fight against the resistance, we propose two perspectives:
- First, identify the resistance genes in order to propose new molecules with different mechanisms of action. For example, antifungal molecules derived from extracts of Balanites aegyptiaca which is a plant of the traditional pharmacopeia used in Burkina Faso to treat superficial candidiosis.
- Secondly, avoid self-medication by prescription of antifungals based on the results of antifungal susceptibility.
Acknowledgments
This work was supported by the Department of Parasitology-Mycology of the university hospital Yalgado Ouedraogo. Our thanks go to:
- Dr Issiaka Soulama, Head of the Molecular Biology Laboratory at CNRFP
- Mr Samuel Sermé from the Molecular Biology Laboratory of CNRFP
References
- Diop M. Profils épidémiologique, clinique, biologique, thérapeutique et évolutif des candidoses oropharingées chez les personnes vivant avec le VIH au centre hospitalier universitaire Yalgado Ouédraogo [Thèse méd]. [Ouagadougou]: Université ouaga 1 Pr Joseph KI-ZERBO, Unité de formation et de recherche en science de santé(UFR/SDS); 2014.
- Bhattacharya S, Sobel JD, White TC. A Combination Fluorescence Assay Demonstrates Increased Efflux Pump Activity as a Resistance Mechanism in Azole-Resistant Vaginal Candida albicans Isolates. Antimicrob Agents Chemother. 2016; 60(10): 5858?66.
- Ford CB, Funt JM, Abbey D, et al. The evolution of drug resistance in clinical isolates of Candida albicans. eLife. 2015; 4: 56-62.
- Li X, Hou Y, Yue L, et al. Potential Targets for Antifungal Drug Discovery Based on Growth and Virulence in Candida albicans. J Antimicrob Chemother. 2015; 59(10): 5885?91.
- Morio F, Pagniez F, Besse M, et al. Deciphering azole resistance mechanisms with a focus on transcription factor-encoding genes TAC1, MRR1 and UPC2 in a set of fluconazole-resistant clinical isolates of Candida albicans. Int J Antimicrob Agents. 2013; 42(5): 410?5.
- Zida A, Issiaka S, Sanata B, et al. 25S rDNA genotype and antifungal susceptibility of clinical Candida albicans in Ouagadougou (Burkina Faso). J Mycol Med. 2017; 8: 101-109.
- Chneider S, Morschhäuser J. Induction of Candida albicans Drug Resistance Genes by Hybrid Zinc Cluster Transcription Factors. Antimicrob Agents Chemother. 2015; 59(1): 558?69.
- Jia C, Zhang K, Yu Q, et al. Tfp1 is required for ion homeostasis, fluconazole resistance and N-Acetylglucosamine utilization in Candida albicans. Biochim Biophys Acta BBA - Mol Cell Res. 2015; 1853(10): 2731?44.
- Poulain D. Candida albicans, plasticité et pathogénie. Rev Francoph Lab. mars 2013; 2013(450): 37?46.
- Ramírez-Zavala B, Mogavero S, Schöller E, et al. SAGA/ADA Complex Subunit Ada2 Is Required for Cap1- but Not Mrr1-Mediated Upregulation of the Candida albicans Multidrug Efflux Pump MDR1. Antimicrob Agents Chemother. 2014; 58(9): 5102?10.
- Sitterlé E, Coste A, Maufrais C, et al. Analyse du polymorphisme des gènes de Candida albicans impliqués dans la résistance aux azolés et aux échinocandines et description de potentielles nouvelles mutations de résistance aux azolés chez des isolats cliniques. J Mycol Médicale. 2017; 27(3): e14?5.
- Sun X, Lu H, Jiang Y, et al. CaIPF19998 Reduces Drug Susceptibility by Enhancing the Ability of Biofilm Formation and Regulating Redox Homeostasis in Candida albicans. Curr Microbiol. 2013; 67(3): 322?6.
- Whaley SG, Tsao S, Weber S, et al. The RTA3 Gene, Encoding a Putative Lipid Translocase, Influences the Susceptibility of Candida albicans to Fluconazole. Antimicrob Agents Chemother. 2016; 60(10): 6060?6.
- Arati M, Pallavi V, Chanchal K, et al. : Molecular mechanisms associated with Fluconazole resistance in clinical Candida albicans isolates from India. Mycoses. 2016; 59: 93–100.
- Jensen RH, Astvad KMT, Silva LV, et al. Stepwise emergence of azole, echinocandin and amphotericin B multidrug resistance in vivo in Candida alcans orchestrated by multiple genetic alterations. J Antimicrob Chemother. 2015; 70(9): 2551?5.
- Katarzyna JSC, ?lemp-Migiel A, Rother M, et al. Nucleotide substitutions in the Candida albicans ERG11 gene of azole-susceptible and azole-resistant clinical isolates. Acta Biochim Pol. 2013; 60: 547–552.
- Xiang MJ, Liu JY, Ni PH, et al. Erg11 mutations associated with azole resistance in clinical isolates of Candida albicans. FEMS Yeast Res. 2013; 13(4): 386?93.
- Caban M, Strapagiel D, Ma?gorzata JD, et al. Principles of a New Protocol for Prediction of Azole Resistance in Candida albicans Infections on the Basis of ERG11 Polymorphisms. Curr Microbiol, 2016; 16: 139-43.
- Go??bek K, Strzelczyk JK, Owczarek A, et al. Selected mechanisms of molecular resistance of Candida albicans to azole drugs. Acta Biochim Pol. 2015; 62(2): 247?51.
- Xu Y, Sheng F, Zhao J, et al. ERG11 mutations and expression of resistance genes in fluconazole-resistant Candida albicans isolates. Arch Microbiol. 2015; 197(9): 1087?93.
- Carvalho VO, Del Negro GMB. Is the S405F mutation in Candida albicans ERG11 gene sufficient to confer resistance to fluconazole? J Mycol Médicale. sept 2014; 24(3): 241?2.
- Ying Y, Zhao Y, Hu X, et al. In Vitro Fluconazole Susceptibility of 1,903 Clinical Isolates of Candida albicans and the Identification of ERG11 Mutations. Microb Drug Resist. 2013; 19 (4): 266?73.
- Feng W, Yang J, Xi Z, et al. Mutations and/or Overexpressions of ERG4 and ERG11 Genes in Clinical Azoles-Resistant Isolates of Candida albicans. 2017 Microb Drug Resist. 2017; 23(5): 563-570.
- Eddouzi J, Parker JE, Vale-Silva LA, et al. Molecular Mechanisms of Drug Resistance in Clinical Candida Species Isolated from Tunisian Hospitals. Antimicrob Agents Chemother. 2013; 57(7): 3182?93.
- Liu JY, Shi C, Wang Y, et al. Mechanisms of azole resistance in Candida albicans clinical isolates from Shanghai, China. Res Microbiol. 2015; 166(3): 153?61.
- Liu Z, Myers LC. Mediator Tail Module Is Required for Tac1-Activated CDR1 Expression and Azole Resistance in Candida albicans. J Antimicrob Chemother. 2017; 61(11): 1342-17.
- Alvarez-Rueda N, Fleury A, Logé C, et al. The amino acid substitution N136Y in Candida albicans sterol 14alpha-demethylase is involved in fluconazole resistance. JMed Mycol. 2016; 54(7): 764?75.
- Hu L, Du X, Li T, et al. Genetic and phenotypic characterization of Candida albicans strains isolated from infectious disease patients in Shanghai. J Med Microbiol. 2015; 64(Pt_1): 74?83.
- Dudiuk C, Gamarra S, Jimenez-Ortigosa C, et al. Quick Detection of FKS1 Mutations Responsible for Clinical Echinocandin Resistance in Candida albicans. Warnock DW, éditeur. J Clin Microbiol. 2015; 53(7): 2037?41.
- Arendrup MC. Multidrug-Resistant Candida: Epidemiology, Molecular Mechanisms, and Treatment. J Infect Dis. 2017; 216: 445–451.
- Carvalho VO, Okay TS, Melhem MSC, et al. The new mutation L321F in Candida albicans ERG11 gene may be associated with fluconazole resistance. Rev Iberoam Micol. 2013; 30(3): 209?12.
- Dannaoui É. Résistance des Candida aux antifongiques?: détection et mécanismes. Rev Francoph Lab. 2013; 2013(450): 71?7.
- Wang H, Kong F, Sorrell TC, et al. Rapid detection of ERG11 gene mutations in clinical Candida albicans isolates with reduced susceptibility to fluconazole by rolling circle amplification and DNA sequencing. BMC Microbiol. 2009; 9(1): 167.
- Zhao J, Xu Y, Li C. Association of T916C (Y257H) mutation in Candida albicans ERG11 with fluconazole resistance: T916C mutation in Candida albicans ERG11 and fluconazole resistance. Mycoses. 2013; 56(3): 315?20.
- Zhang L, Yang HF, Liu YY, et al. Reduced susceptibility of Candida albicans clinical isolates to azoles and detection of mutations in the ERG11 gene. Diagn Microbiol Infect Dis. 2013; 77(4): 327?9.
- Salari S, Khosravi AR, Mousavi SAA, et al. Mechanisms of resistance to fluconazole in Candida albicans clinical isolates from Iranian HIV-infected patients with oropharyngeal candidiasis. J Med Mycol. 2016; 26(1): 35-41.
- Strzelczyk JK, Slemp-Migiel A, Rother M, et al. Nucleotide substitutions in the Candida albicans ERG11 gene of azole-susceptible and azole-resistant clinical isolates. Acta Biochim Pol. 2013; 60(4): 547?52.
- Li X, Cai Q, Mei H, et al. The Rpd3/Hda1 family of histone deacetylases regulates azole resistance in Candida albicans. J Antimicrob Chemother [Internet]. 29 mars 2015 [cité 5 avr 2018]; Disponible sur: https://academic.oup.com/jac/article-lookup/doi/10.1093/jac/dkv070
- Ren B, Dai HQ, Pei G, et al. ABC transporters coupled with the elevated ergosterol contents contribute to the azole resistance and amphotericin B susceptibility. Appl Microbiol Biotechnol. 2014; 98(6): 2609?16.
- Popp C, Hampe IAI, Hertlein T, et al. Competitive Fitness of Fluconazole-Resistant Clinical Candida albicans Strains. Antimicrob Agents Chemother. 2017; 61(7): 584-17.
- Srikantha T, Daniels KJ, Pujol C, et al. Identification of Genes Upregulated by the Transcription Factor Bcr1 That Are Involved in Impermeability, Impenetrability, and Drug Resistance of Candida albicans a/ α Biofilms. Eukaryot Cell. 2013; 12(6): 875?88.
- Lackner M, Tscherner M, Schaller M, et al. Positions and Numbers of FKS Mutations in Candida albicans Selectively Influence In Vitro and In Vivo Susceptibilities to Echinocandin Treatment. Antimicrob Agents Chemother. 2014; 58(7): 3626?35.
- Suwunnakorn S, Wakabayashi H, Rustchenko E. Chromosome 5 of human pathogen Candida albicans carries multiple genes for negative control of caspofungin and anidulafungin susceptibility. Antimicrob Agents Chemother. 2016; AAC.01888?16.
- Wang Y, Liu JY, Shi C, et al. Mutations in transcription factor Mrr2p contribute to fluconazole resistance in clinical isolates of Candida albicans. Int J Antimicrob Agents. 2015; 46(5): 552?9.
- Vincent BM, Lancaster AK, Scherz-Shouval R, et al. Fitness Trade-offs Restrict the Evolution of Resistance to Amphotericin B. PLoS Biol. 2013; 11(10): e1001692.
